Studies at the Intersection of Ribosome Function and Cellular Homeostasis
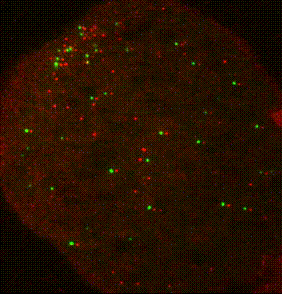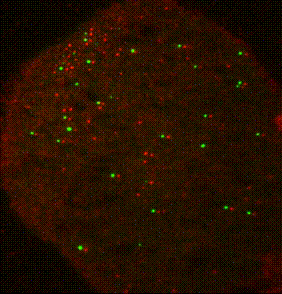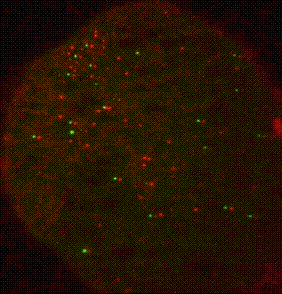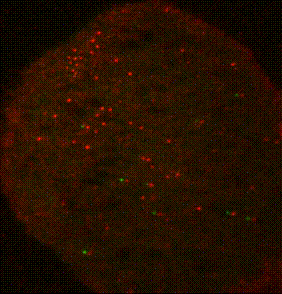
The ribosome is a complex molecular machine that translates the genetic code into functional polypeptides. Our work focuses on understanding how the ribosome functions at a molecular level and how changes in its activity lead to mRNA quality control and the induction of cellular stress responses. Work in the Green lab ranges widely in scope, from detailed mechanistic questions in ribosome rescue to surveying global changes in gene expression and dissecting the complex interplay of mammalian signaling pathways. We use a wide range of genetic, genomic, and biochemical approaches to explore these questions in bacteria, yeast, and increasingly in mammalian systems. Much of our ongoing work focuses on the importance of ribosome collisions in activating signaling pathways such as the integrated stress response (ISR) and various MAP kinase cascades.
Watch Rachel Green’s iBiology.org video explaining Protein Synthesis at https://www.ibiology.org/biochemistry/protein-synthesis/"